A 56-year-old Japanese man had spent 2 months in Freetown, Sierra Leone, for medical work. He had been prescribed doxycycline for malaria prophylaxis, but occasionally missed doses and discontinued it upon returning to Japan. He had not traveled abroad for at least 6 months before the onset of symptoms. Two months after his return, he developed recurrent nocturnal fevers that resolved spontaneously the following day. These episodes recurred five times at intervals of 12–48 h. On the 10th day after symptom onset, he presented to the hospital.
Upon arrival at the hospital, his body temperature was 37.9 °C, blood pressure 137/72 mmHg, and heart rate 95 bpm. Physical examination revealed no signs of jaundice in his ocular conjunctiva or hepatosplenomegaly. Blood tests showed the following: red blood cell count 4.72 × 105/µL, hemoglobin 13.5 g/dL, platelet count 11.0 × 104/µL, aspartate aminotransferase 34 IU/L, alanine aminotransferase 64 IU/L, and lactate dehydrogenase 311 IU/L. There was no evidence of hypoglycemia, metabolic acidosis, renal impairment, or hyperbilirubinemia. Additionally, a chest X-ray showed no abnormalities. Blood cultures showed no growth.
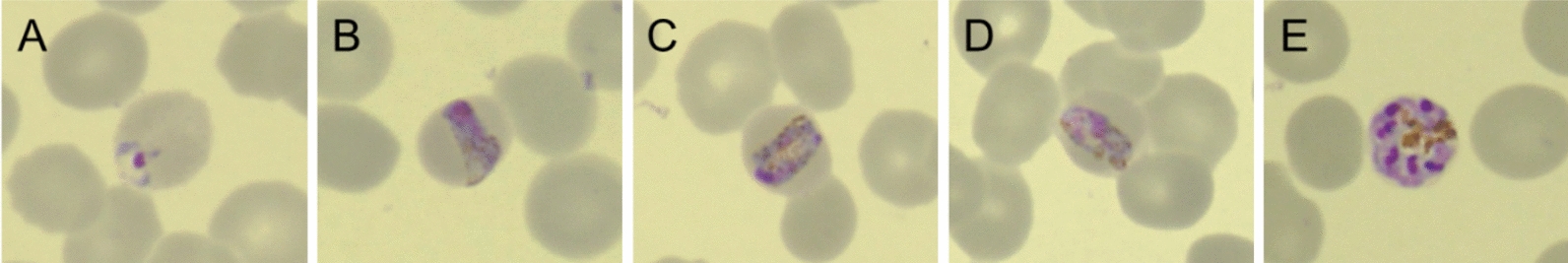
A rapid diagnostic test (BinaxNow Malaria; Abbott) indicated non-falciparum malaria. Microscopic observation of Giemsa-stained thin-blood smears revealed parasitized erythrocytes with band-shaped trophozoites, characteristic of P. malariae, with a parasitemia of 0.059% (27.8 × 102/µL) (Fig. 1).
Microscopic images of Giemsa-stained thin-blood smears (× 1000 magnification). A Ring form; B–D band form; E schizont
The patient was diagnosed with uncomplicated quartan malaria and treated with artemether–lumefantrine for 3 days. He was discharged without complications after a fever clearance time of approximately 27 h and a parasite clearance time of approximately 36 h. These clearance times were within the expected range for artemether–lumefantrine treatment, indicating a favorable therapeutic response. He remained asymptomatic for 4 months after discharge.
Nested PCR was positive only for P. malariae, confirming species identification [1,2,3]. However, agarose gel electrophoresis showed that the secondary PCR band using P. malariae-specific primers was fainter than the primary PCR band obtained with universal primers (Fig. 2). Sequence analysis of the SSU rRNA gene revealed a four-base deletion at the binding site of the species-specific reverse primer (Fig. 3). Cytochrome b gene sequencing confirmed 100% identity with P. malariae (Fig. 4) [4].
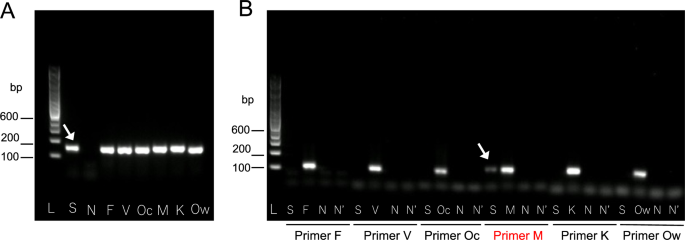
Results of nested polymerase chain reaction (PCR). A Primary PCR. Lane L: 100 bp DNA ladder. Lanes S, F, V, Oc, M, K, and Ow: the arrow indicates DNA templates from the patient’s blood (S) and positive controls containing partial small subunit ribosomal ribonucleic acid (SSU rRNA) gene sequences of Plasmodium species, including P. falciparum (F), P. vivax (V), P. ovale curtisi (Oc), P. malariae (M), P. knowlesi (K), and P. ovale wallikeri (Ow). Lane N: negative control (water); B secondary PCR. Lane L: 100 bp DNA ladder. Lanes S, F, V, Oc, M, K, and Ow: first-round PCR products using the same templates as in panel A. Lane N: first-round product from water. Lane N′: water-only control. The arrow indicates the PCR product amplified from the patient’s sample using P. malariae-specific primers (S). Primer sets used for each species are indicated below the gel

Partial sequence analysis of the small subunit ribosomal ribonucleic acid (SSU rRNA or 18S rRNA) (317 bp). A total of 372 base pairs (bp) were amplified using the originally designed forward primer (P0.5: CTGCGTTTGAATACTACAGCATGGA), located upstream of the primer used in the primary PCR (P1: ACGATCAGATACCGTCGTAATCTT [1, 2], shown in blue), and the reverse primer from the primary PCR (P2: GAACCCAAAGACTTTGATTTTTCTCAT [1, 2]). Of these, 317 bp were successfully sequenced. The 18S rRNA sequence of the present case was deposited to DDBJ/EMBL/GenBank under the accession number LC859598. This sequence, labeled as “Patient”, was aligned with the complete SSU rRNA sequence of Plasmodium malariae, labeled as “Pm SSU rRNA” (DDBJ/EMBL/GenBank accession number XR_003751927.1). In the patient’s DNA sequence, four bases corresponding to the P. malariae-specific reverse primer (indicated in red), used in nested PCR, were found to be deleted

Partial sequence analysis of the Cytochrome b gene (357 bp). A 400-bp fragment amplified using two primers (RTPCRSc2_F: TGGAGTGGATGGTGTTTTAGA and Sc3R: ACCCTAAAGGATTTGTGCTACC [4]) yielded 357 bp of readable sequence. The data were deposited in DDBJ/EMBL/GenBank under the accession number LC859864. This sequence, labeled as “Patient”, was aligned with the complete mitochondrial DNA sequence of Plasmodium malariae, labeled as “Pm mit.” (DDBJ/EMBL/GenBank accession number LT594637.1). The patient’s sequence was 100% identical to the cytochrome b gene of P. malariae