This study was carried out within the framework of EDCTP (European & Developing Countries Clinical Trials Partnership)-granted projects DIAGMAL (Phase 3 evaluation of an innovative simple molecular test for the diagnosis of malaria in different endemic and health settings in sub- Saharan Africa, RIA2018D-2496) and e-MANIC (Development and evaluation of an electronic algorithm using a combination of a two-step malaria RDT and other rapid diagnostic tools for the management of febrile illness in children under 5 attending outpatient facilities in Burkina Faso,TMA2019CDF-2697) aiming respectively to: (i) evaluate the field performance of the mini-dbPCR-NALFIA test for the diagnosis of malaria in different clinical and malaria settings, and (ii) to tackle antimicrobial resistance at primary health centres by implementing point-of-care (PoC) tests, including malaria sequential diagnostic with PfHRP2 and pLDH [13, 14]. Briefly, all febrile patients (presenting with an axillary temperature ≥ 37.5 °C or with a history of fever during the previous seven days), regardless of age, attending the outpatient clinic of the selected health facilities were invited to participate in the study. For seasonality purpose, monthly attendance for fever of the previous year was used to distribute the sample size for one-year recruitment covering the range of the two malaria transmission seasons in Burkina Faso (Supplement Table 1). Signed informed consent was obtained from participants, and parents/guardians for minors prior to data and clinical specimen collection. Demographic data, medical history and the history of use of antimalarial drugs within the past four weeks were recorded for each participant. At recruitment site, participants were screened with malaria RDT detecting PfHRP2 recommended by the Ministry of Health for malaria diagnosis in rural areas and, if positive, the participants were managed according to the national guidelines. For the purpose of the study, a dried blood spot (DBS) was subsequently made by collecting finger prick blood on Whatman 3 filter paper. A capillary blood (200 µL) was collected in an ethylene diamine tetra acetic acid (EDTA) tube for expert microscopy, dbPCR-NALFIA and two-steps malaria RDTs (PfHRP2/pLDH) testing.
Both the DIAGMAL and e-MANIC studies have been approved by the National Ethics Committee for Health Research of Burkina Faso (DIAGMAL: deliberation Nº2021-03–057, 10 March 2021; e-MANIC: deliberation Nº2021-04–084, 07 April 2021).
Laboratory procedures
Microscopy
Thick and thin blood smears for microscopy were prepared in duplicate from capillary blood collected in EDTA tubes and stained with Giemsa 3% for 30 min. At CRUN (Clinical Research Unit of Nanoro), a double-reading system is routinely used to ensure accurate and reliable results. To further enhance quality, CRUN collaborates with the National Institute for Communicable Diseases (NICD) in South Africa as part of an external quality assessment programme, demonstrating its commitment to maintaining high standards in diagnostics. In case of discrepancy, a decisive reading was performed by a third expert reader. Discrepancy includes: disagreements on whether the sample was positive or negative, differences in identifying the Plasmodium species, or difference between the higher and lower parasite density exceeding 50% of the lower reading [15, 16]. Microscopists were blinded from the malaria RDT results. A quality control was performed on 10% of slides by independent expert microscopists at Amref Health Africa in Kenya.
Malaria rapid diagnostic test
All consented participants were tested systematically for malaria using the Ministry of Health’ PfHRP2-based RDT (SD Bioline Pf: Standard Diagnostic, Hagal-Dong, Republic of Korea). Routine health centre staff performed the test and recorded the result on the study case report form (CRF).
Two-steps malaria RDT detecting both PfHRP2 and pLDH were performed using capillary blood collected in EDTA tube by trained laboratory technicians according to the manufacturer’s instruction (SD Bioline® Malaria Ag P.f/Pan batches 05EDG030A, 05EDG063A: Standard Diagnostics, Republic of Korea). The test was valid if the internal control line was positive. Results were recorded separately for each band test (pLDH and PfHRP2) as positive or negative after 15–30 min. In case of discrepancy between two independent readers, the opinion of a third independent reader was requested. The result was recorded for each antigen.
dbPCR-NALFIA test
This assay is a duplex reaction based on the detection and amplification of two regions Plasmodium 18S rRNA genes: one that is highly conserved and specific for pan-Plasmodium, and a second that is specific to P. falciparum [17, 18]. The test procedure consist of five main steps, as previously described [12, 19]: (i) preparation of the master mix; (ii) lysis of the whole blood; (iii) adding the dbPCR mix to the lysed blood; (iv) dbPCR run; and, (v) NALFIA test. The running time of the dbPCR is estimated at 80 min. The dbPCR-NAFIA strip is read after 10 min of migration. For all NALFIA test performed, a picture was taken and saved in secured computer.
Performance study of the sequential interpretation of PfHRP2 and pLDH combined with dbPCR-NALFIA
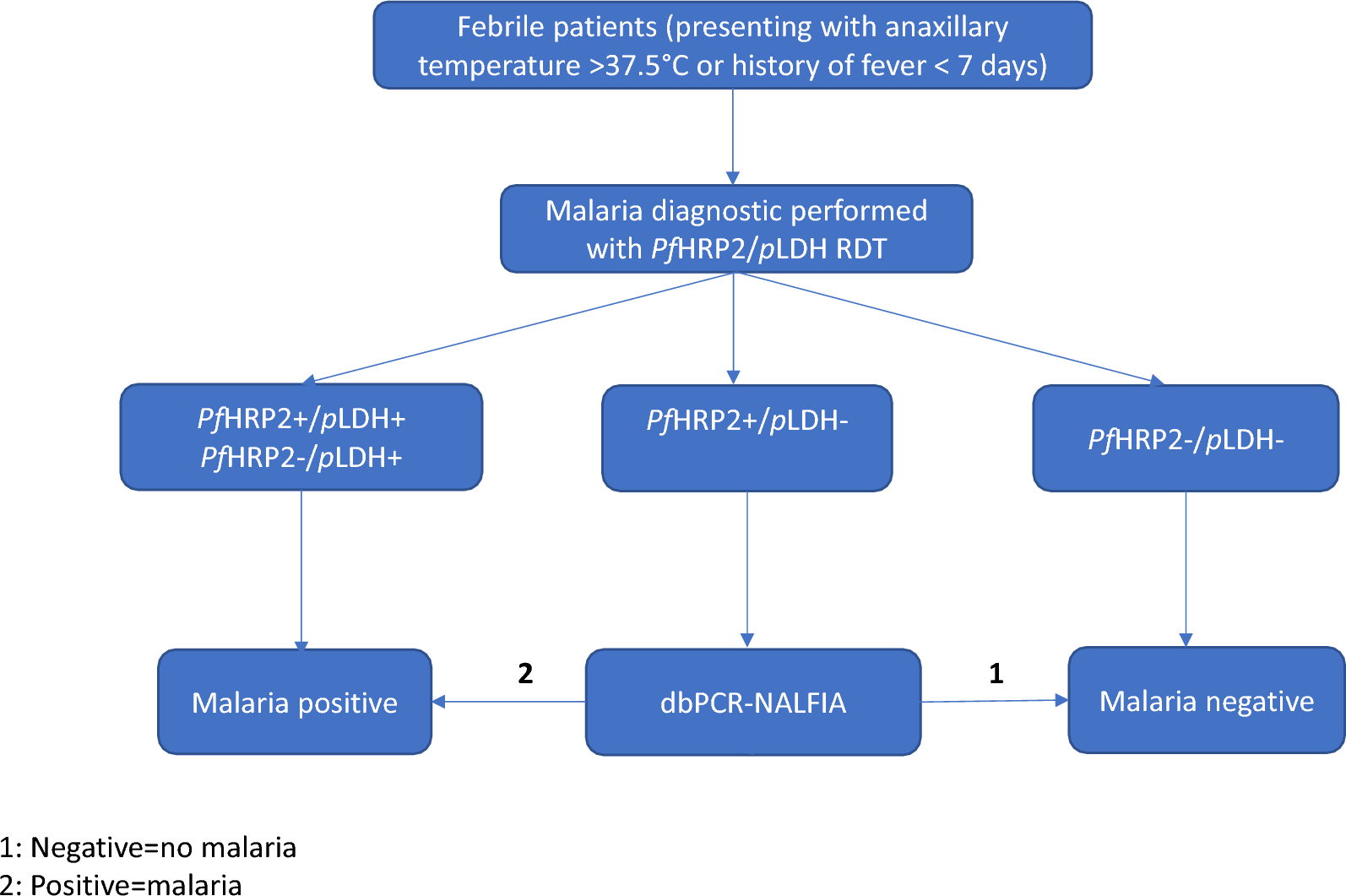
To assess the performance of the proposed algorithm combining PfHRP2 and pLDH, with dbPCR-NALFIA, the diagnostic results of the PfHRP2 and pLDH antigens were recorded as well as the dbPCR-NALFIA results. The interpretation of the two-step malaria RDT detecting PfHRP2 and pLDH and the mini-dbPCR-NALFIA was done as follows:
-
PfHRP2(+)/pLDH(+): falciparum malaria or co-infection with non-falciparum malaria;
-
PfHRP2(−)/pLDH(+): non-falciparum malaria or falciparum malaria with deletion of hrp2;
-
PfHRP2(−)/pLDH(−): negative results
-
PfHRP2(+)/pLDH(−): inconclusive results and dbPCR-NALFIA is needed to differentiate:
-
If dbPCR-NALFIA is positive (both P. falciparum and Pan test lines or only Pan test line appear), the malaria diagnosis is reported as positive result. The positive test result will be recorded has P. falciparum or Pan.
-
If dbPCR-NALFIA is negative (only the test control line appear), the malaria diagnosis is reported as negative result.
The proposed sequential algorithm workflow is summarized in Fig. 1.
Sequential algorithm workflow combining the two-step malaria RDT detection PfHRP2/pLDH and dbPCR-NALFIA
Deoxyribonucleic acid (DNA) extraction and VarATS quantitative PCR
This qPCR was performed in case of: (i) discordance between microscopy and RDT (both SD Bioline Pf, SD Bioline® Malaria Ag P.f/Pan), and (ii) undetermined diagnostic results for SD Bioline® Malaria Ag P.f/Pan.
Genomic DNA was extracted from DBS using the QIAamp DNA mini kit® (Qiagen, Germany) according to the manufacturer’s recommendations and stored at − 20 °C. Five (5) µL of DNA were used as template for qPCR analysis targeting P. falciparum var gene acidic terminal sequence (varATS, ≈59 copies per genome) as previously described [20]. qPCR was run on StepOnePlus Real-Time PCR (Applied Biosystems™, Thermo Fisher Scientific, Waltham, MA, USA). Parasite densities were determined by interpolating cycle thresholds (Ct) using a standard curve prepared with titrated samples (100,000–0.1 parasites/μL). The limit of detection of the varATS-based qPCR was 0.4 parasite/μL for DNA extracted from filter paper. Samples with Ct value > 38.0 were considered negative. DNA extracted from P. falciparum-positive sample with high parasite density obtained from patients not included in this study was used as positive control. The negative controls included human negative blood spots on filter paper and master mix reagent used as negative template control (NTC).
Data collection and analysis
Data were double-entered using OpenClinica software. Statistical analysis was performed with STATA software version 17 (StataCorp, College Station, Texas 77,845, USA). Description of qualitative and quantitative variables were performed by using proportion with 95% confidence intervals (CI) and mean ± SD [or median with interquartile (IQR) ranges], respectively. The geometric mean was used to express the parasite densities. The performance of sequential algorithm was evaluated compared to standard diagnosis test in place (PfHRP2-RDT) by calculating the sensitivity, the specificity, negative and positive predictive values with the microscopy as gold standard on conclusive diagnostic results. The qPCR was used as gold standard for undetermined results tested with dbPCR-NALFIA. The Cohen’s Kappa values were calculated to measure the level of agreement between sequential algorithm and reference tests (microscopy and qPCR). The Kappa value used in interpretation was calculated according to the standard classification proposed by McHugh [21]. p ≤ 0.05 was considered statistically significant.