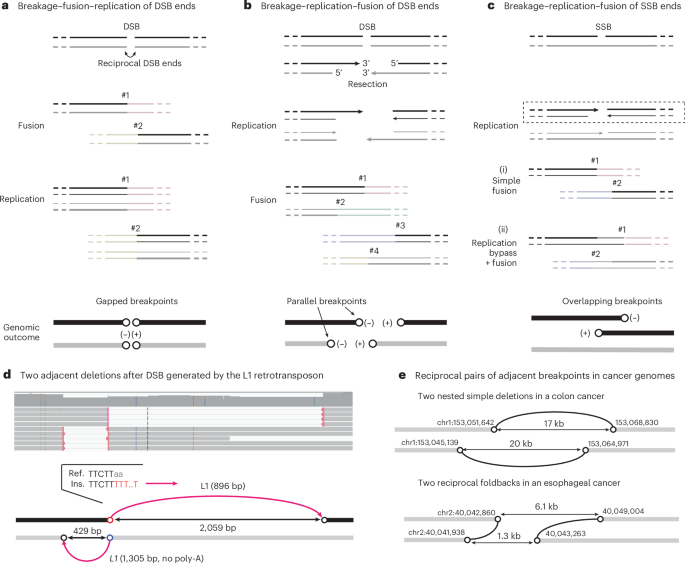
We first show how breakage–replication/fusion converts free DNA ends into breakpoints on rearranged sequences and then show how breakage–replication/fusion of chromosome fragments produces segmental copy-number gains and amplifications. We place particular emphasis on distinguishing the genomic feature of a rearranged DNA sequence (for example, breakpoints) from the molecular feature of the ancestral chromosome (for example, DNA ends). See Supplementary Note, Section 1 for the complete list of definitions.
Rearrangements from breakage–replication/fusion of DNA ends
A DNA double-strand break (DSB) generates two reciprocal DNA ends (Fig. 1a). In the G1 phase, these ends can undergo classical non-homologous end-joining (c-NHEJ): they can be ligated together, creating a rearranged sequence with small deletions (or less frequently, duplications), or ligated to DNA ends from distal sites, creating translocations29,30. In either scenario, the ancestral DNA ends are converted to two breakpoints (open circles in Fig. 1a) separated by a small gap, which we term adjacent gapped breakpoints. As the ligation(s) occur before replication, the rearranged DNA sequences are preserved in both sister chromatids after replication. This cascade of events defines the breakage–fusion–replication sequence (Supplementary Video 2).
If the DSB ends have substantial overhangs that prevent c-NHEJ31,32 (for example, because of 5′-resection33,34,35 or 3′-exonuclease degradation24), they can remain unligated during G1 and persist into S phase. During S phase, these ends, like broken chromosome ends36, are replicated to generate two ‘sister’ DNA ends. Ligations of these replicated DNA ends can generate up to four rearrangement junctions (Fig. 1b). This cascade of events defines the breakage–replication–fusion sequence (Supplementary Video 3). In breakage–replication–fusion, a staggered DNA end is converted into two adjacent but non-identical breakpoints with the same orientation, which we term adjacent parallel breakpoints. When the sister DNA ends are directly ligated to each other, it produces a ‘foldback’ junction, joining two adjacent parallel breakpoints. Foldback junctions are often assumed to indicate fusions between the ends of broken sister chromatids in BFB cycles37,38; later, we will show that such fusions also occur between sister DNA fragments.
In a variation of breakage–replication–fusion, two single-strand DNA (ssDNA) ends with a small gap are converted into two reciprocal DSB ends by replication39,40 (Fig. 1c). These two DSB ends can generate two rearrangement breakpoints with either a small gap (i) or a small overlap (ii) by a replication bypass mechanism18,41,42. We refer to the latter as adjacent overlapping breakpoints.
A single DSB end undergoes either breakage–fusion–replication or breakage–replication–fusion. However, when a catastrophic event creates many DNA breaks, some will undergo breakage–replication–fusion while others will undergo breakage–fusion–replication; we refer to the latter as the breakage–replication/fusion cycle.
Adjacent parallel breakpoints from DNA end replication
We first sought experimental evidence that a single DNA end can generate two adjacent parallel breakpoints. We exploited L1 retrotransposition to simultaneously generate and mark DSB ends. As described in a separate paper43, transient L1 expression in p53-null RPE-1 cells generated both L1 insertions and translocation junctions containing reverse-transcribed L1 sequences. Both outcomes originate from DSB ends generated by the L1 open reading frame 2 protein (ORF2p), and are identified by the insertion of reverse-transcribed sequences (the ‘primary’ end of retrotransposition) and/or the presence of ORF2p endonuclease target sequences near the break site.
We identified multiple instances of adjacent parallel breakpoints in clones generated after L1 induction that had features indicating an origin from ORF2p-induced DSBs (Supplementary Note, Section 6). In the example shown in Fig. 1d, two nested deletions, each containing a truncated L1 insertion, indicate two pairs of adjacent parallel breakpoints (Fig. 1b). The sequence features at the two closest breakpoints (red and blue circles) directly relate them to L1 ORF2p, and the distances between each pair of parallel breakpoints (429 bp and 2,059 bp) are consistent with DSB resection33,34,35. Together, these observations demonstrate that breakage–replication–fusion can generate two parallel breakpoints from a single DSB end.
Footprints of DNA end replication in human disease genomes
We next sought evidence of DSB end replication in human disease genomes. Although we cannot directly relate a rearrangement breakpoint to an ancestral DNA end, we can identify an ancestral DSB from breakpoints derived from reciprocal DSB ends: in particular, a reciprocal pair of parallel breakpoints directly identifies reciprocal DSB ends that undergo breakage–replication–fusion (Fig. 1b,d).
Based on the observations from the L1 clones (Fig. 1d and Supplementary Note, Section 6), we selected a heuristic threshold distance of 20 kb for the identification of adjacent parallel breakpoints (Methods). From 592,176 breakpoints detected in 2,588 cancers by the Pan-Cancer Analyses of Whole Genomes (PCAWG) study16, we identified 20,795 pairs of adjacent parallel breakpoints from 1,793 samples. These breakpoints were identified at 35,422 rearrangement junctions (12% of all junctions), including 7,393 foldback junctions. Thus, adjacent parallel breakpoints are a widespread feature in cancer genomes.
For 3,138 pairs of adjacent parallel breakpoints, we identified one or multiple reciprocal breakpoints that demonstrate their origin from ancestral DSBs. There were 417 instances of reciprocal parallel breakpoints as shown in Fig. 1b. Among these were 53 instances of nested deletions (Supplementary Table 1) and 23 instances of reciprocal foldbacks (Supplementary Table 2), with examples shown in Fig. 1e. In the remaining instances, one or multiple breakpoints formed long-range translocations. Examples of reciprocal foldbacks were previously noted in ovarian cancers (supplementary fig. 8f of a previous publication44) but were assumed to result from BFB cycles. We suggest that these events arise from reciprocal DSB ends undergoing breakage–replication–fusion.
We further assessed the probability that adjacent parallel breakpoints were generated independently based on the distance between these breakpoints and their distance to the nearest breakpoint on the opposite side (Methods). This analysis showed that for 16,132 of 20,795 pairs of adjacent parallel breakpoints, the probability that they were generated independently was less than 5%.
In summary, adjacent parallel breakpoints are common in cancer genomes, and our analysis suggests that many of them are derived from sister DNA ends generated by breakage–replication–fusion.
DNA duplication and amplification from breakage–replication/fusion
When the sister DNA ends are joined together in a single rearranged chromosome, this rearranged chromosome will contain a duplication (Fig. 2a). Moreover, the duplicated segments will be bounded by adjacent parallel breakpoints derived from sister DNA ends. Consistent with this prediction, we identified examples of copy-number gains flanked by adjacent parallel breakpoints in both human cancers and congenital diseases14,15,45,46 (Extended Data Fig. 1 and Supplementary Note, Section 7).
Foldback junctions are the simplest outcome when sister DNA ends are joined together. We envision two processes by which a double-stranded DNA (dsDNA) fragment can generate amplification with only foldback junctions (Fig. 2b). If both ends of a dsDNA fragment undergo breakage–replication–fusion to form foldback junctions (Fig. 2b (ii)), the outcome is a dimeric circular DNA (previously termed type II episomes47). Like simple monomeric DNA circles (type I episomes47; Fig. 2b (i)), dimeric DNA circles can fuel DNA amplification by asymmetric segregation over successive generations. This model explains the amplification at the AR locus flanked by foldback junctions in a castration-resistant prostate cancer46 (Extended Data Fig. 1a, right). Amplification can also occur on a linear acentric DNA fragment when the DNA ends on opposite sides fuse asynchronously (Fig. 2b (iii)). If sister DNA ends on one side are fused together, but sister DNA ends on the opposite side remain unligated (red arrows), the product is a linear inverted dimer. In the next cell cycle, another round of replication–fusion can create a circular or linear tetramer without any new breakage. Iterations of this process will produce a large tandem array of amplified DNA with ‘nested’ foldbacks that form homogeneously staining regions of inverted duplications48,49.
One such example is the amplification spanning the ERBB2 oncogene in the HCC1954 breast cancer genome (Fig. 2c). Similar patterns were also found in chr8p, chr12p and chr20q in this genome (Extended Data Fig. 2a–c and Supplementary Tables 3 and 4). Here, amplified ERBB2 is contained in homogeneously staining regions37,50 and is bounded by multiple foldback junctions previously attributed to BFB cycles37. However, the probability of generating foldback junctions in such close proximity by successive BFB cycles is very small (see Fig. 2c caption). Under the breakage–replication/fusion model, the close proximity between foldback junctions near 39.5 Mb is a natural consequence of the close proximity between the 3′ and 5′ ends of an ancestral DSB end (Fig. 2b (iii)). Moreover, if amplification takes place on a linear, extra-chromosomal DNA fragment, secondary breakpoints (both foldbacks and long-range breakpoints) can only arise within the amplicon, thus explaining the concentration of breakpoints within the amplified region (39.5–40 Mb). Importantly, in linear DNA amplification, amplified DNA is automatically doubled and linked in one chromosome that is segregated into one daughter cell, thus providing a more rapid route to higher DNA copy number than amplification by random segregation of episomal circles. The amplification of DNA copy number also does not require selection during the intermediate steps of amplification; therefore, focal amplifications lacking oncogenes (Extended Data Fig. 2a–c) may be passengers that undergo clonal fixation.
In summary, the presence of duplicated or amplified DNA segments flanked by adjacent parallel breakpoints suggests an origin from breakage–replication/fusion. From a single acentric DNA fragment, breakage–replication/fusion can generate dimeric DNA circles or a linear array of inverted duplications with closely spaced foldbacks, explaining the long-standing observation of inverted duplications in amplified DNA47,48,49 that are unlikely to arise by multi-generational BFB cycles37,38,44.
Segmental copy-number gains after chromosome fragmentation
Above, we described the rearrangement and copy-number outcomes of breakage–replication/fusion occurring at a single dsDNA end and a single dsDNA fragment with two ends. Below, we describe the copy-number and rearrangement outcomes of breakage–replication/fusion after chromosome fragmentation.
We focused the analysis on an experimental model of chromothripsis (Fig. 3a, left) because this system enabled us to determine the structure of rearranged chromosomes with near-complete resolution (Methods). In a previous study21, we used CRISPR–Cas9 to generate chromosome bridges containing dicentric chr4 and derived single cells with a broken chr4 (Supplementary Note, Section 8). In one generation, bridge breakage produced daughter cells with reciprocal DNA retention and deletion21 similar to what was observed immediately after micronucleation20. However, over many generations, clones derived from single cells frequently had subclonal copy-number gains without reciprocal loss in the sibling clone21. The presence of copy-number gains in clones expanded after chromosome fragmentation was also observed in clones expanded after telomere crisis24 (Supplementary Note, Section 9) or micronucleation25 (Extended Data Fig. 3 and Supplementary Table 5).
One bridge clone (primary clone 1a from a previous publication21, hereafter referred to as clone a) is interesting because the bulk DNA copy number oscillates between variable non-integer states that indicate subclonal copy-number variation (Fig. 3a, middle). Moreover, some subclones showed two-state copy number oscillation while others showed segmental copy-number gains (Fig. 3a, right, and Fig. 3b; also see fig. S22 from previous work21). The presence of subclonal copy-number variation enabled us to first determine the breakpoints of duplicated segments and then infer the evolutionary history of the rearrangements that produced the duplications (Methods and Supplementary Note, Sections 10 and 11). Based on a joint analysis of segmental DNA copy number (Supplementary Table 8) and rearrangement junctions (Supplementary Table 9), we determined both the structure (Extended Data Fig. 4) and the joining pattern (Extended Data Fig. 5) of nearly all duplicated segments in the subclones of clone a. In total, we identified 86 rearranged segments with sizes above 10 kb (Supplementary Tables 10–12) and 126 short insertions (<10 kb) between these segments (Supplementary Tables 13–16). We next show that the genomic features of the large segments, the short insertions and their arrangement in the rearranged chromosomes indicate that they all originate from breakage–replication/fusion of a single chromatid.
Large duplications from breakage–replication/fusion
The origin of large duplications in clone a from ancestral chromosome fragments that underwent breakage–replication/fusion is established by two orthogonal lines of evidence that relate the boundaries of the duplications to ancestral DNA ends.
First, we identified 18 pairs of duplicated segments that are flanked by identical (‘flush’) or adjacent parallel (‘staggered’) breakpoints within 20 kb (Fig. 4). Knowing the exact size of each duplicated segment, we could assess the probability that the staggered breakpoints were generated independently using the ratio of breakpoint distance to the segmental size (Extended Data Fig. 6). Based on this metric, we determined that for 15 out of 20 pairs of staggered breakpoints, the probability of independent breakpoint generation was less than 0.05 (Supplementary Table 11). For the remaining five pairs, the breakpoint distances were within a similar range but the segments were shorter; therefore, all staggered breakpoints are consistent with an origin from the replication of (hyper)resected DSB ends. For three pairs of segments (Bb1/Bb2, Cb1/Cb2, Cc1/Cc2), the presence of reciprocal breakpoints directly established their origin from chromosome fragmentation (Extended Data Fig. 7a). These data provide statistical evidence that the staggered boundaries of duplicated segments arose from breakage–replication–fusion.
Second, we observed strand-coordinated base substitutions near the staggered breakpoints that directly established their origin from staggered DSB ends. Based on the breakage–replication/fusion model, the shorter breakpoint derives from an ancestral 5′ end and the longer breakpoint derives from an ancestral 3′ end. Thus, the offset region between the two breakpoints originates from the ancestral ssDNA overhang. We identified seven clusters of substitutions near the staggered breakpoints (Fig. 4, downward or upward arrows), six of which were restricted to the offset region (the only exception near the right side of the shorter Bb2 segment is explained in Extended Data Fig. 7b.) All the substitutions reflect deamination in the TpC context that is consistent with the outcome of ssDNA deamination by APOBEC enzymes51. Importantly, the signature of substitutions (C > X on the right side of each segment, downward arrows; G > X on the left side of each segment, upward arrows) directly established the deaminated ssDNA to be a 3′ overhang. Thus, the pattern of deamination between staggered breakpoints provides molecular evidence for their origin from staggered DSB ends. Additional evidence linking staggered breakpoints to staggered DSB ends comes from the coordination between breakpoints on opposite sides of duplicated segments (Extended Data Fig. 7c and caption).
Based on adjacent parallel breakpoints, we determined that 40 duplicated segments in clone a were derived from ancestral sister DNA fragments generated by breakage–replication/fusion (Supplementary Table 10).
DNA over-replication from breakage–replication/fusion
In addition to nearly identical duplications generated by normal, semi-conservative replication of ancestral chromosome fragments, we also identified rare examples reflecting two mechanisms of DNA over-replication. The replication bypass mechanism41,42 (Fig. 1c) explains two instances of overlapping duplications18,52 (Extended Data Fig. 8a–c and caption); the second mechanism, leading to re-replication of a previously replicated DNA fragment, occurs when the previously replicated segment is fused to an unreplicated segment with unfired origins (Extended Data Fig. 8d and caption).
Short insertions from breakage–replication/fusion
We identified 126 short insertions (median size, 184 bp) at the junctions between large duplications in clone a (Supplementary Tables 13–16). Three pieces of evidence indicate that both the insertions and the insertion rearrangement junctions are generated by chromosome breakage–replication/fusion.
First, when mapped to their origin sites, the insertions displayed several features indicating DNA fragmentation. Nearly all insertions (113 out of 126) were mapped to sites in close proximity (<10 kb) to breakpoints inferred to have been derived from ancestral DNA ends. Moreover, at several sites, the insertions lined up one after another in a tiling pattern, with little gap or overlap (Fig. 5a,b). The tiling pattern of insertions at the origin sites is incompatible with random polymerase template-switching events in MMBIR that are expected to generate duplicated sequences with either large gaps or large overlaps at their original sites (Supplementary Video 4). Finally, seven tiles of insertions were mapped right next to breakpoints derived from the 5′ ends of ancestral DSBs (Figs. 4 and 5b and Supplementary Table 13). Similar patterns were also observed in other experimentally generated clones with chromothripsis (Supplementary Note, Sections 6, 9 and 14), in cancer genomes (Extended Data Fig. 9 and Supplementary Note, Section 7) and in congenital disorders53. Based on these observations, we suggest that many insertions originate as ssDNA fragments complementary to the 3′ overhang of resected DSBs. Two potential models for the generation of these insertions are discussed in Supplementary Note, Section 3.
Second, the joining pattern of insertions in rearranged DNA suggested DNA end-joining repair. A total of 111 out of 126 insertions were assembled into 17 chains (c1–c17) of two or more tandem insertions at rearrangement junctions (Supplementary Table 15), 13 of which are shown in Fig. 5c. These chains were only identified at junctions inferred to be breakage–replication–fusion junctions formed in S/G2, but not breakage–fusion–replication junctions formed in G1. Moreover, the junctions between the neighbor insertions within each chain often showed either >2 bp microhomology or additions of non-templated nucleotides. By contrast, breakage–fusion–replication junctions had few insertions and little microhomology, consistent with c-NHEJ in G1. Therefore, the insertion junctions probably arise from microhomology-mediated end-joining of sister DNA ends in breakage–replication–fusion.
Finally, and most definitively, the strand orientation of insertions at their destination junctions suggests that they were incorporated into both DNA strands and could not have arisen from a conservative replicative process14,16 such as MMBIR. Under the MMBIR model11,14, insertions at each junction are continuously added to the 3′ end of the nascent leading strand that jumps from one template to the next; therefore, all the insertions would be added to a single strand in the rearranged DNA. As the original DNA strands of the insertions could be inferred from the adjacency between the insertions and nearby breakpoints (left-facing or right-facing arrows Fig. 5b), we were able to directly test whether the insertions were added to the same strand in the rearranged DNA. If we consider every pair of insertions that are next to each other in every insertion chain (Supplementary Table 15), 38 pairs are added to the same strand (arrows pointing to the same direction in Fig. 5c) but 41 pairs are added to opposite strands (arrows pointing to opposite directions). This observation therefore excludes MMBIR as the mechanism for generating the insertion junctions.
In summary, the genomic features of short insertions in clone a indicate that both the inserted sequences and the insertion junctions were generated in the same breakage–replication/fusion cycle that produced large duplications.
Genomic complexity from one breakage–replication/fusion cycle
Based on the general assumption that breakpoints in close proximity arise at approximately the same time16, we inferred that all the breakpoints and junctions in the ancestral rearranged chr4 of clone a (Extended Data Fig. 5c) were generated in a single breakage–replication/fusion cycle. Moreover, except for the rare instances of over-replication (Extended Data Fig. 8), all the ancestral segments, including short insertions, could be traced to non-overlapping ssDNA fragments. Therefore, the ancestral rearranged chr4 of clone a was most likely derived from a single ancestral chromatid over one breakage–replication/fusion cycle.
Breakage–replication/fusion explains genomic complexity
A single breakage–replication/fusion cycle can generate both segmental duplications flanked by adjacent parallel breakpoints and rearrangement junctions containing insertions originating from DSB ends (Fig. 6). To assess the contribution of breakage–replication/fusion to insertion rearrangements in cancer genomes, we analyzed insertions in the PCAWG data. We identified 85,684 potential insertions with a median size of ~2 kb (Methods and Extended Data Fig. 10a). These insertions accounted for 29% of all rearrangement breakpoints; 48% of insertions (41,445 out of 85,684) were mapped to regions within 10 kb from another breakpoint, but overlapping breakpoints were rare (<5% of insertions show 10 bp or larger overlap). These observations were consistent with the features of insertions generated by the breakage–replication/fusion mechanism (Fig. 5). Moreover, the two signatures of breakage–replication/fusion—adjacent parallel breakpoints and short insertions from a single DSB end—provide intuitive explanations for many complex rearrangement footprints that were identified in the PCAWG study16 but to date had no mechanistic interpretation (Fig. 6b and Extended Data Fig. 10).