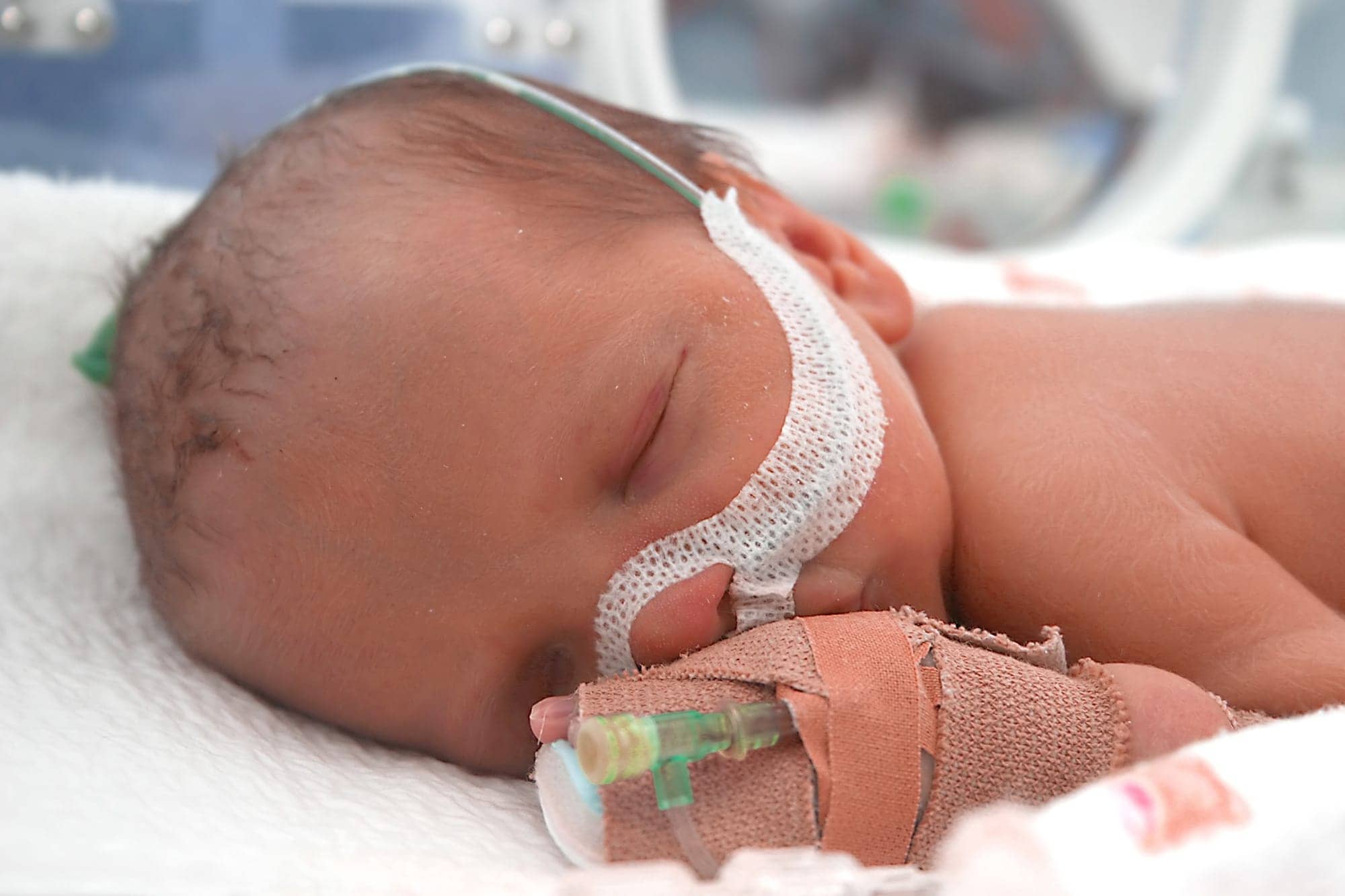
Newborns who receive surgery for a severe form of spina bifida are more likely to suffer from breathing disruptions during sleep than previously known.
RT’s Three Key Takeaways:
- Early sleep-disordered breathing: More than…

A long-term study published in Science Advances has found that while a ketogenic diet limited weight gain in mice, prolonged adherence triggered metabolic disruptions, including impaired insulin secretion, fatty liver disease and elevated blood…

The use of antiviral treatments in children hospitalized for influenza fell sharply during the COVID-19 pandemic, according to a study published last week in Pediatrics.
While 48% to 57% of hospitalized children…
Researchers don’t have a definitive answer on what’s behind young people developing colorectal cancer, but the evidence right now points to lifestyle and environmental factors.
Another factor, he told Healthline, is that the colon is uniquely…

Terms
While we only use edited and approved content for Azthena
answers, it may on occasions provide incorrect responses.
Please confirm any data provided with the related suppliers or
…

In the context of the Data Analysis and Real World Interrogation Network (DARWIN EU®), researchers in the DARWIN EU® Coordination Centre analysed electronic health records from five European countries: Belgium, Germany, the Netherlands, Spain,…